The gwasglue2 package sits between the data and the analytical methods within the OpenGWAS ecosystem
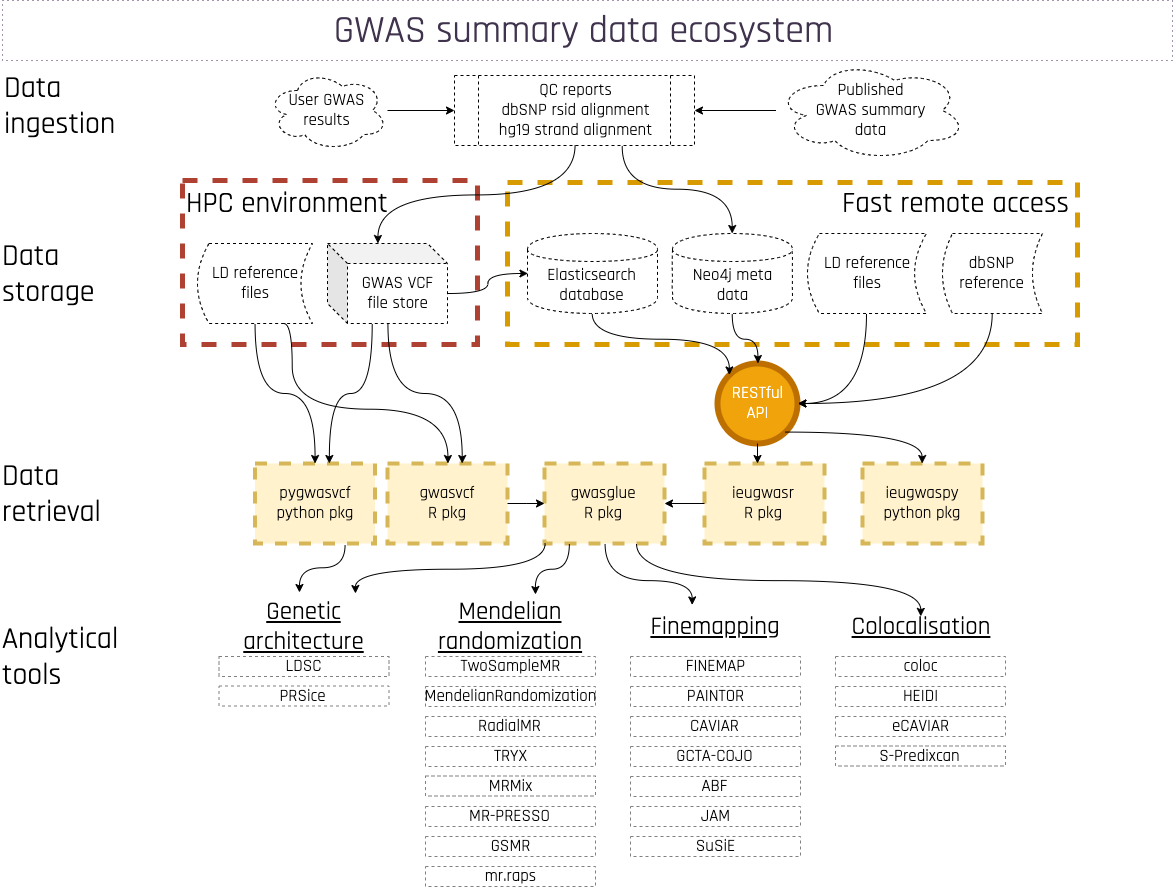
Figure 1: gwas ecosystem
Objectives
- Represent GWAS summary data from multiple sources in various different shapes for different tools
- Represent a class structure that other packages can call
- Have very few dependencies
- Ensure harmonisation of data across different sources
- Be able to simulate data for ease of analytical prototyping
- Store relevant meta data
- Have methods for visualising, joining and subsetting datasets
- Create examples of analytical tools creating methods that import the gwasglue2 package.
Here are some example ‘structures’ of summary datasets that it should be able to handle
Data structures

Figure 2: Data structures
Note that the whole OpenGWAS database is an example of a ‘complete’ dataset - all traits and all variants. What we typically want to represent in gwasglue2 is a specific slice of that data, that is almost always going to be a rectangular shape - a set of variants across a set of traits. In addition, we need to be able to be able to flexibly associate metadata or other forms of annotations (LD matrices, genomic annotations etc) to the data. Finally, analytical tools need to be aware of these structures in order to easily deploy methods upon them.
Work flow
This is typically how a workflow might look:

Figure 3: Work flow
We should be able to ingest data from various locations, and then the class provides a number of different ways to manipulate and annotate the data.
Strategy
A set of genetic associations (1 or more) for a single trait is known as a SummarySet. This class should contain the data, plus MetaData that describes the source study, plus a list of Concepts that annotate the Summary data.
A Concept is a named list of extra information that can be linked to the data, for example LD matrices, annotations relating to how to use the data in a model (e.g. exposure or outcome), etc.
A Dataset is a set of merged and harmonised SummarySets, for example if two SummarySets are generated using the same set of variants for two traits, then putting them together into a Dataset will find the intersect of the available variants, and harmonise them to be on the same effect allele. Dataset objects can also have a global Concept list for extra data that is common across all the SummarySets stored in the object.
Initial visualisation below:
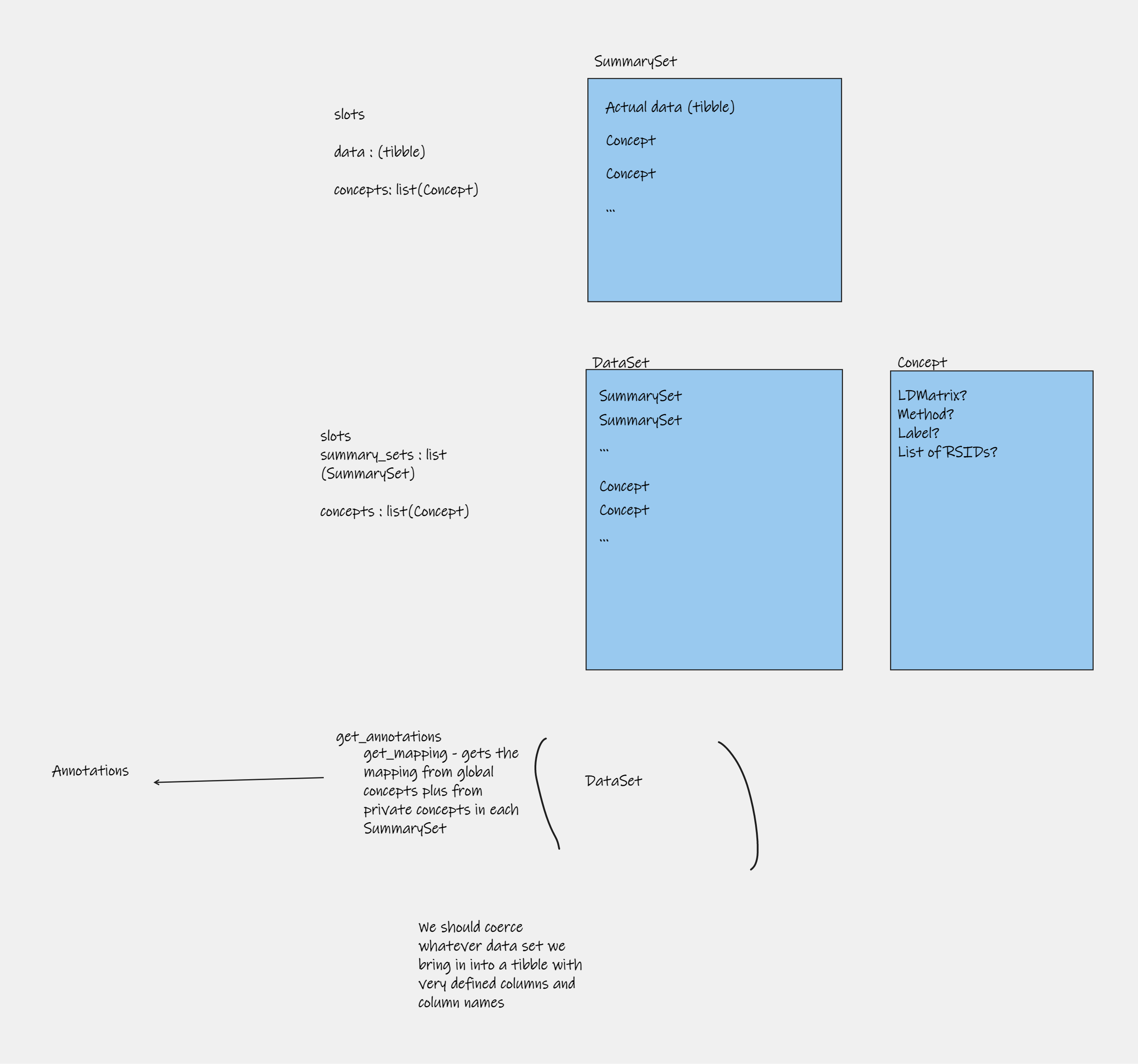
Figure 4: Class structures
Implementation
Currently planning to implement as an S4 class, but may move into R7 in the future once it is stable and compatible with roxygen etc.
Controlled fields
Association data within a SummarySet should have the following fields -
- Required:
- chromosome
- position
- effect allele (ALT)
- non-effect allele (REF)
- effect size
- standard error
- p-value
- Optional:
- effect allele frequency
- rsid
- imputation info score
- sample size
- number of cases
- number of controls
Metadata fields -
- Required
- genome build
- units of effect size
- number of cases
- number of controls
- trait name
- trait identifier
- ancestral population of sample
- sex of sample
- study DOI
- number of variants
- Optional
- Ontology term
- standard deviation of trait
- mean of trait
- genotyping method
- analysis method
- covariates
- notes
Accession info should be generated upon creation:
- Data source
- Timestamp of access
- Query
Design
gwasglue2 has several constructors to build the SummarySet and DataSet objects, as well as getter and setter methods to add or retrieve information within.
To build the SummarySet-class object, we need just one constructor function, the create_summaryset(). It calls different create_summaryset_from_(), depending of the source type of the GWAS summary data and also creates metadata using information from the summary data if none is given. It is also possible to build a metadata object using create_metadata() and input it in create_summaryset(). Fig. 5 shows the structure design of SummarySet with the getters and setters on the left and constructors on the right.
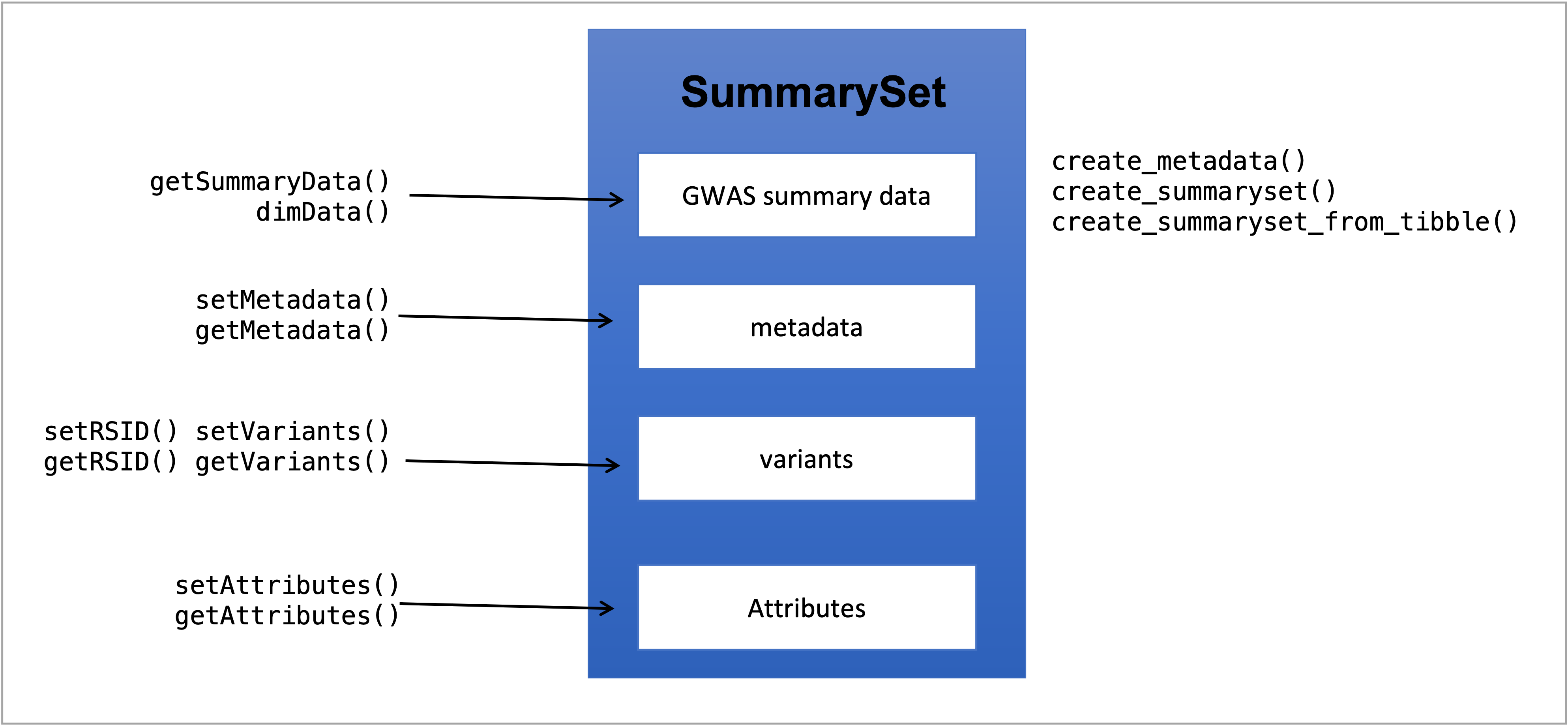
Figure 5: SummarySet
For DataSet-class object we use the create_dataset() constructor. If argument harmonise = TRUE it creates an harmonised DataSet. We use the function harmonise_ld() to harmonise the DataSet against a LD matrix from a reference population. The meta_analysis() function statistically combines two or more SummarySets within the DataSet and creates a new SummarySet object. With add_summaryset() it is possible to add an existent SummarySet to the DataSet and harmonise again. Two or more DataSets can also be merged together with merge_datasets(). The structure design for DataSet is in Fig. 6, with all the getters and setters on the left and constructors on the right.
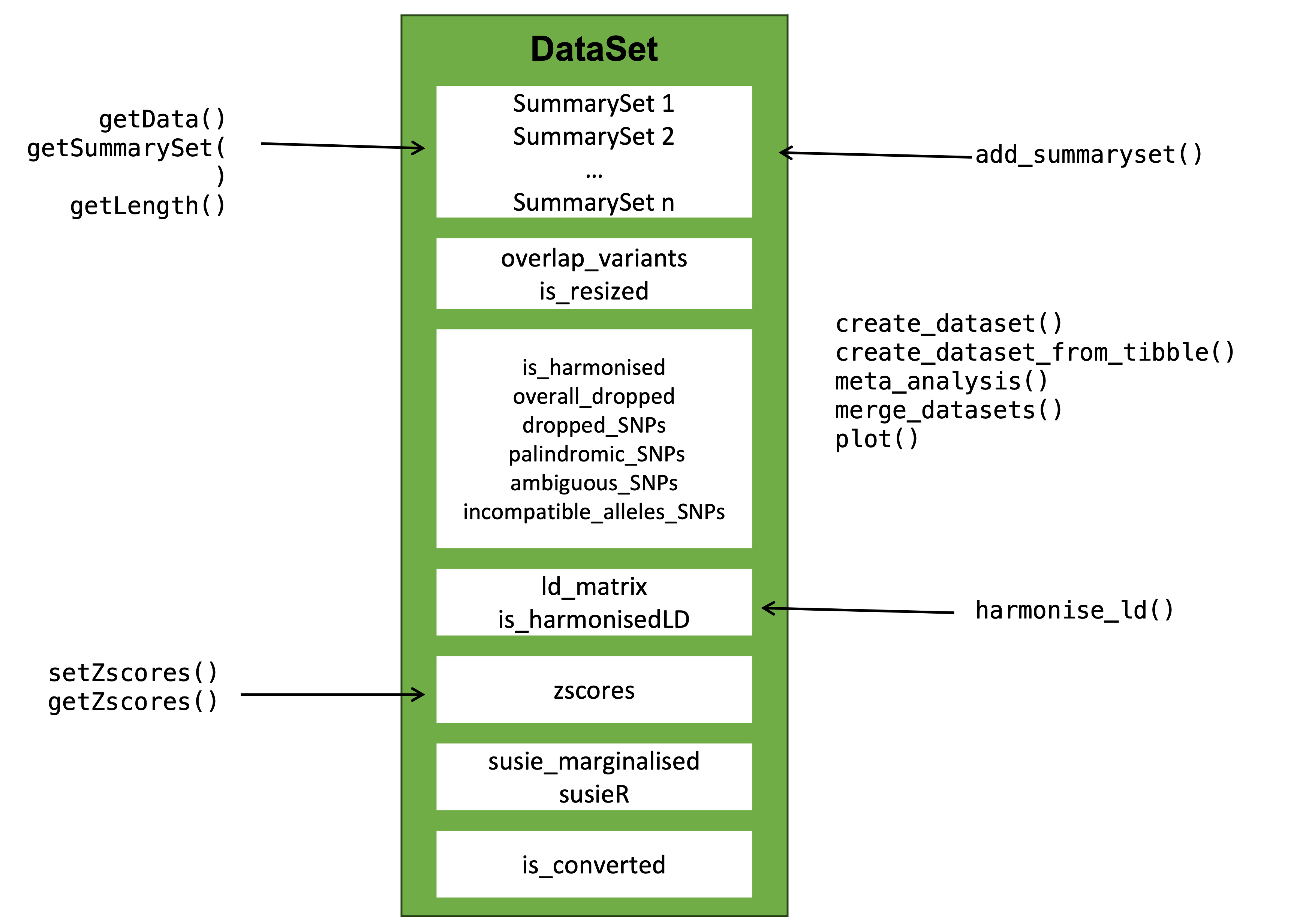
Figure 6: DataSet
Variantid
Opposite to gwasglue, gwasglue2 does not uses rsids when harmonising the data or in other analyses. Instead, a variantid is created using the chromosomal (chr) position (pos) and effect (ea) and non-effect (nea) alleles, taking the form of chr:pos_ea_nea. If the allele has more than 10 characters (e.g. indels), gwasglue2 will hash it using the murmur32 algorithm from the R digest package.
The GWAS summary data is also standardised when creating a SummarySet and transformed for the alleles to be in alphabetical order. Thus, the effect allele will always be the first alphabetically.
| DATA | CHR | POS | EA | NEA | EA Frequency | BETA | variantid |
|---|---|---|---|---|---|---|---|
| raw | 1 | 123 | A | G | 0.3 | 1 | |
| gwasglue2 | 1 | 123 | A | G | 0.3 | 1 | 1:123_A_G |
| raw | 1 | 123 | G | A | 0.3 | 1 | |
| gwasglue2 | 1 | 123 | A | G | 0.7 | -1 | 1:123_A_G |
| raw | 1 | 123 | GTAGTAGTAGTA | A | 0.3 | 1 | |
| gwasglue2 | 1 | 123 | A | GTAGTAGTAGTA | 0.7 | -1 | 1:123_A_#bf37641c |