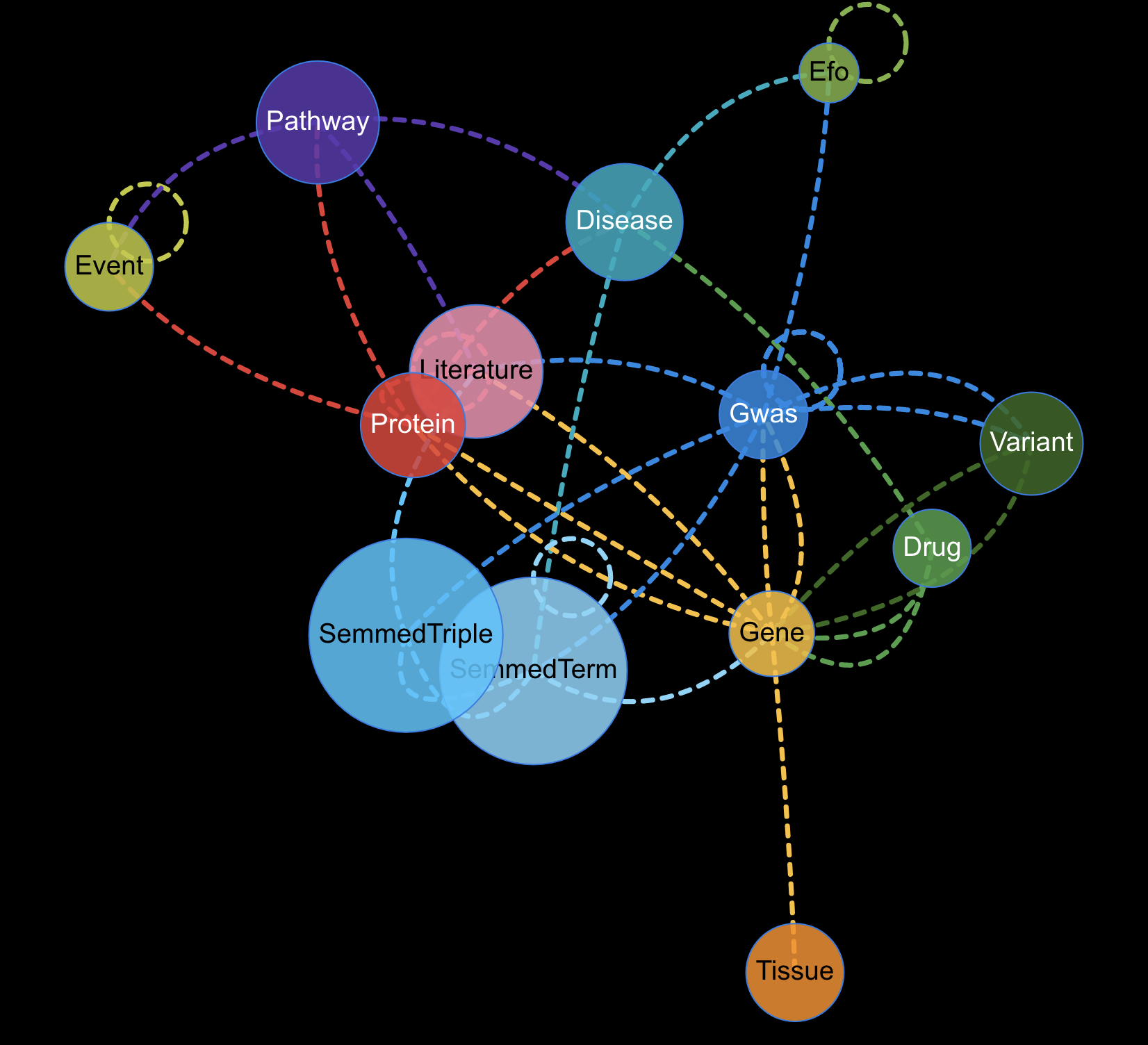
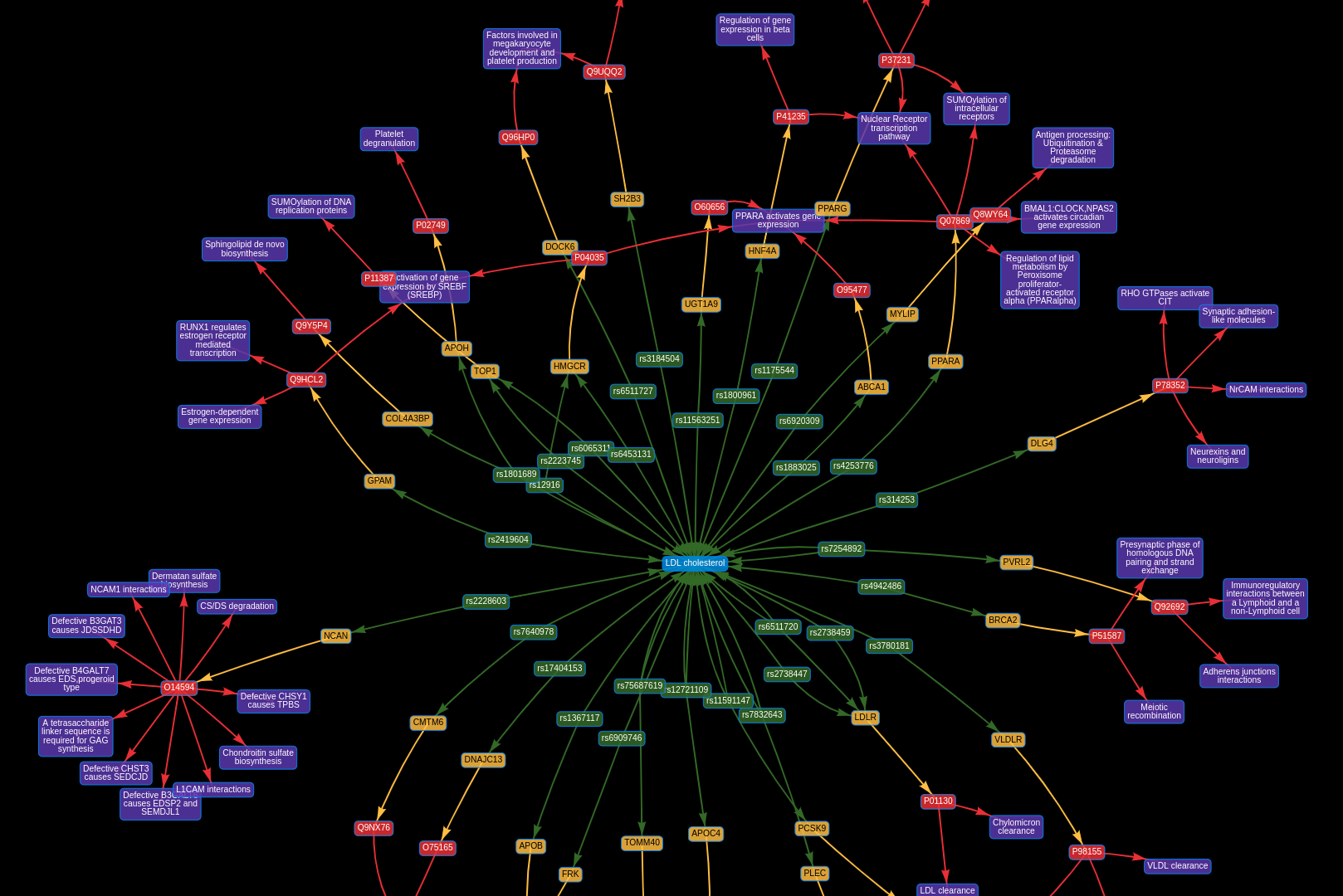
Welcome to MRC Integrative Epidemiology Unit GitHub pages.
The MRC Integrative Epidemiology Unit at the University of Bristol (IEU) conducts some of the UK’s most advanced population health science research. It uses genetics, population data and experimental interventions to look for the underlying causes of chronic disease.
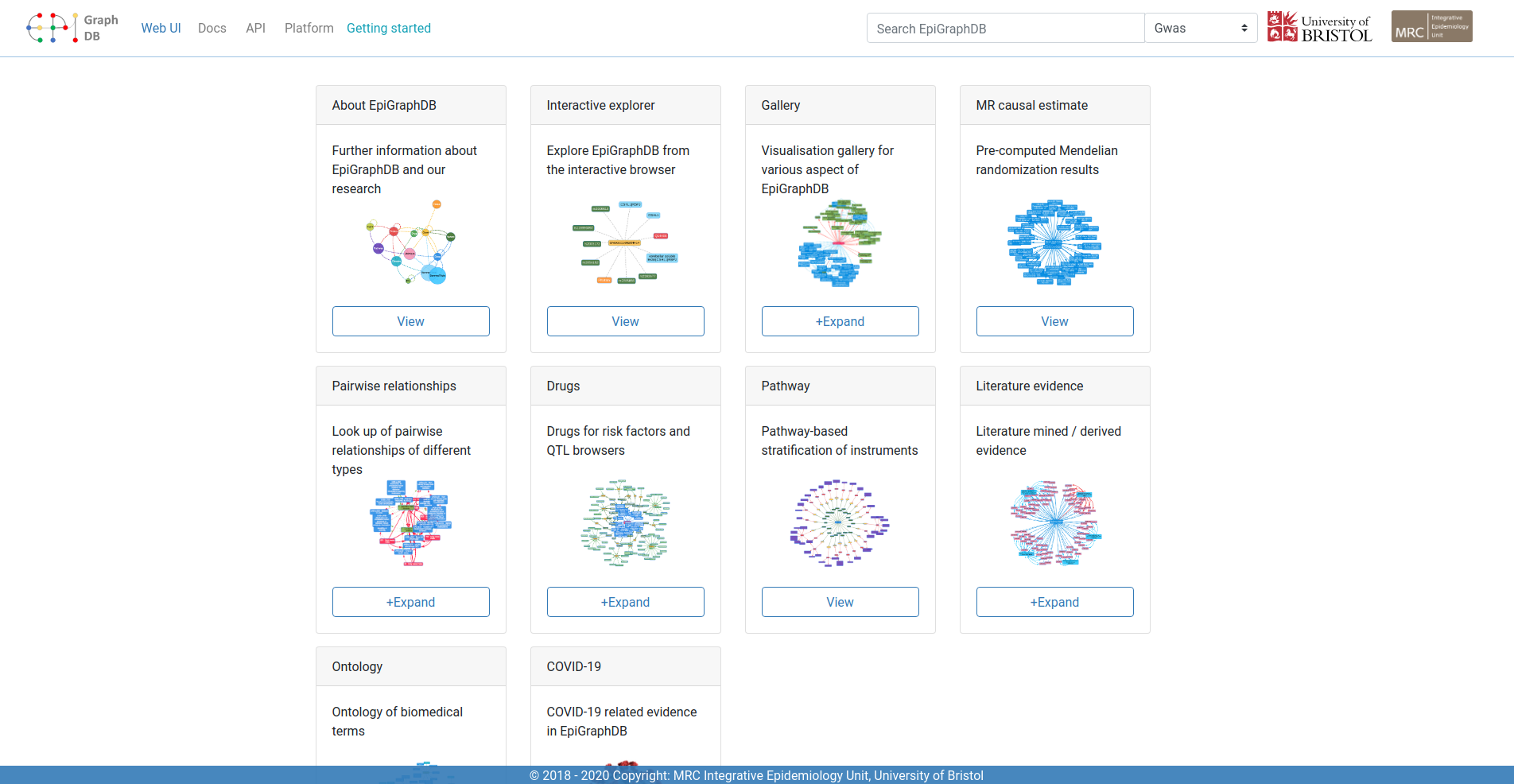
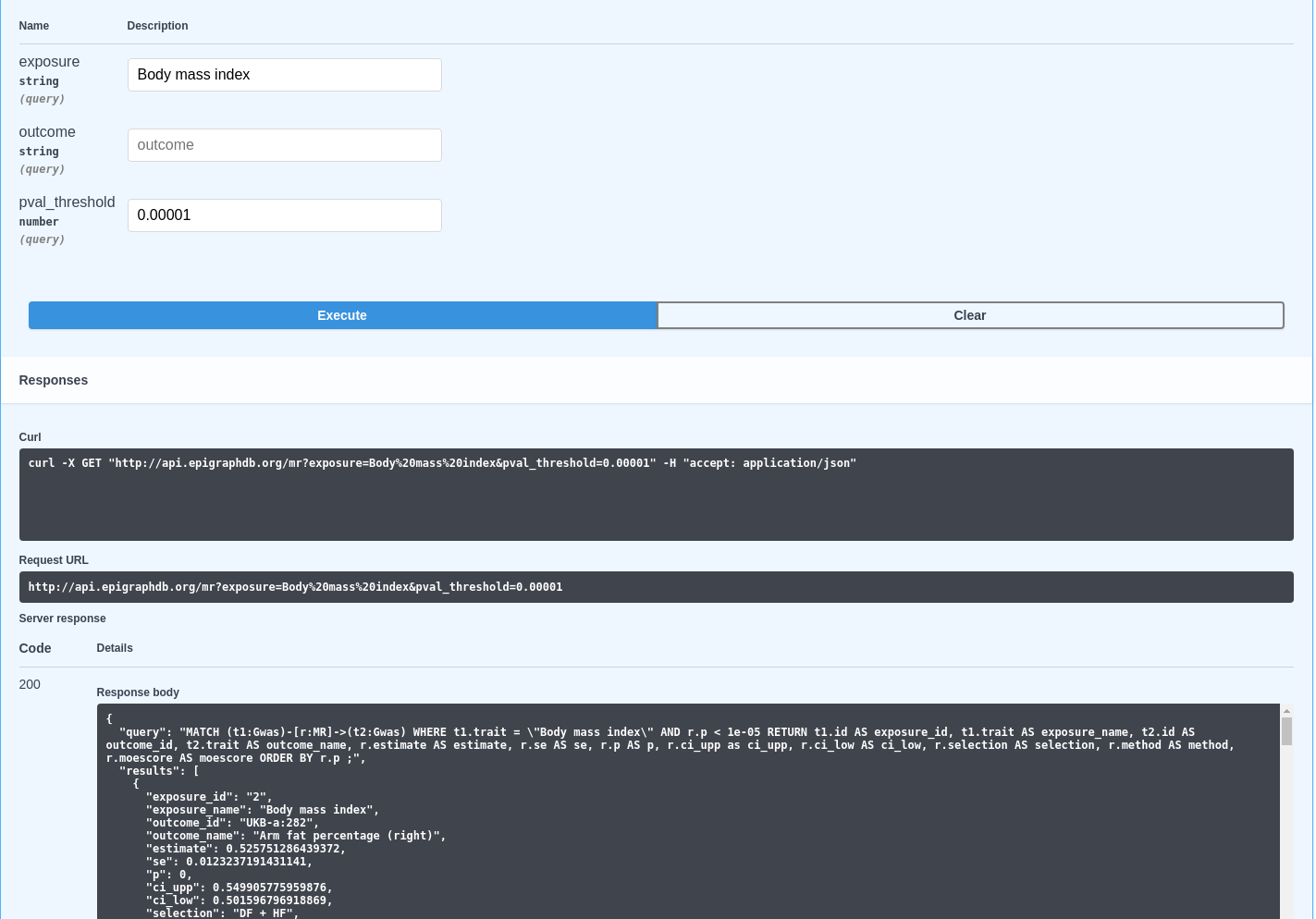
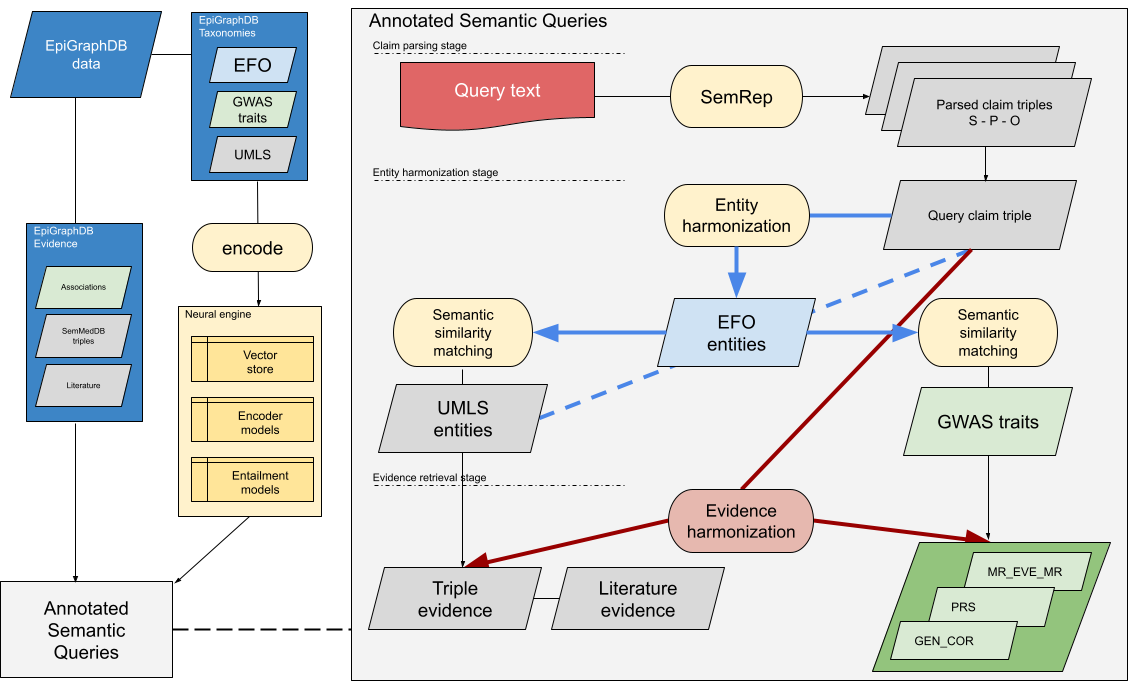
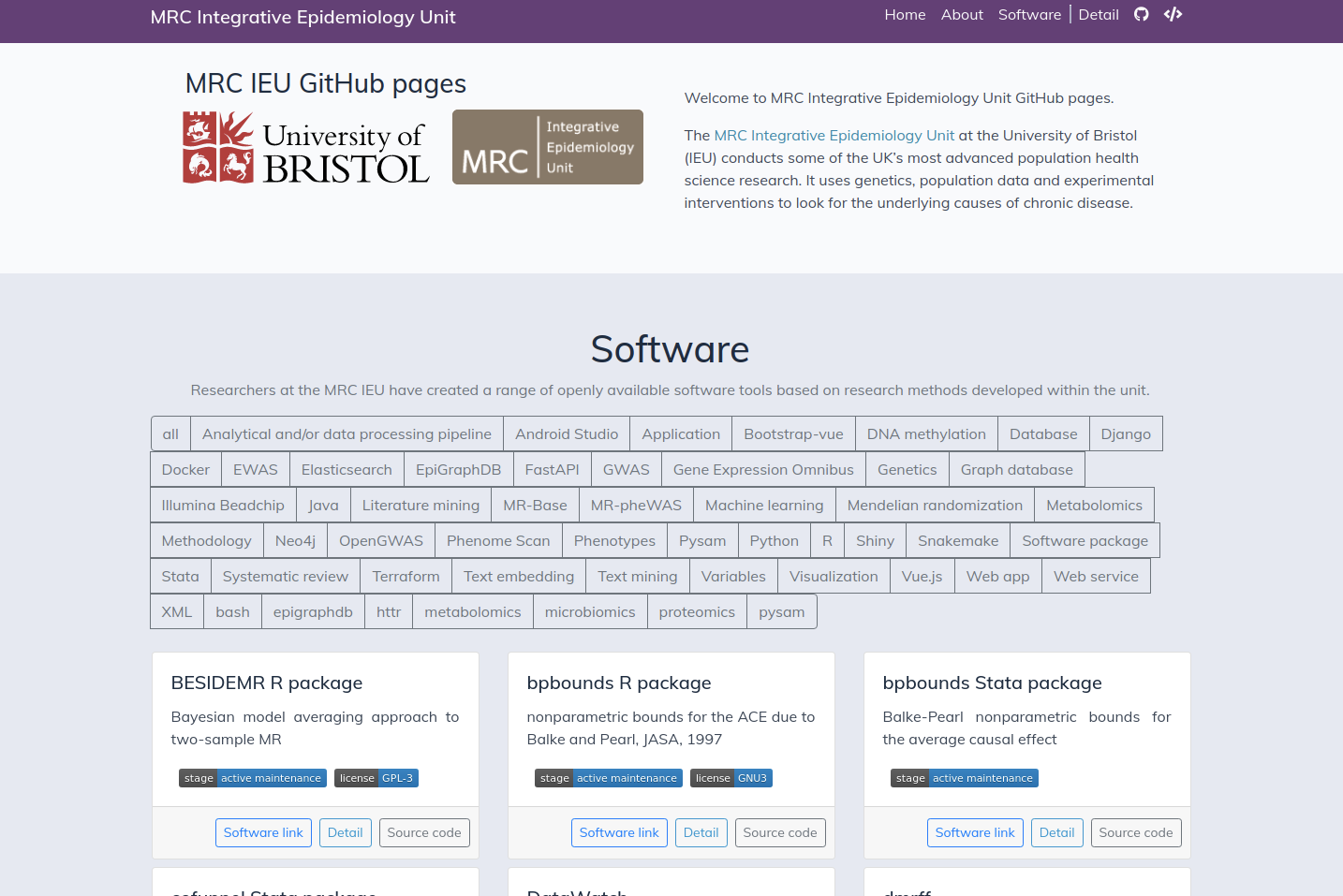
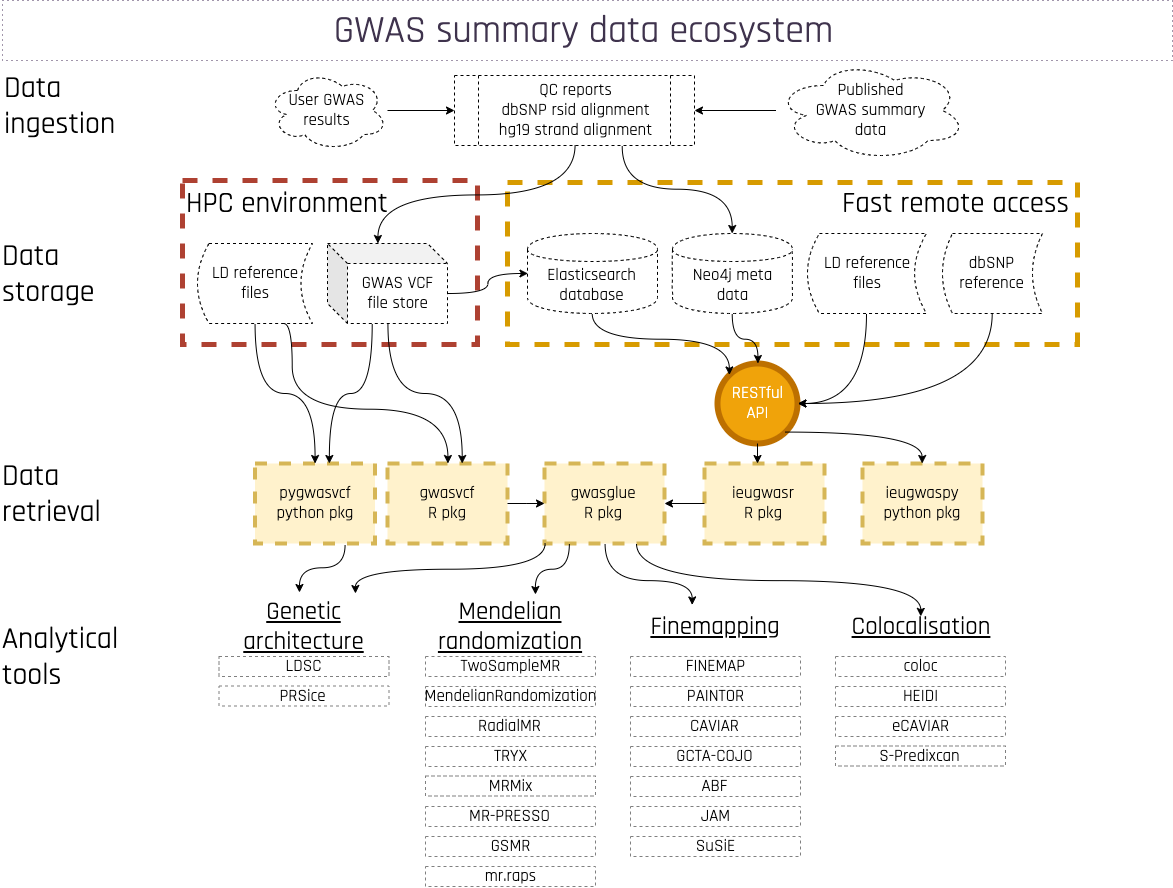
Researchers at the MRC IEU have created a range of openly available software tools based on research methods developed within the unit.